Authors:
Jehan Mahmoud Mahmoud Ouf1, Yuan Yuan2, Prashant Singh2 and Azlin Mustapha2*
1Food Hygiene Department, Animal Health Research Institute, Dokki, Giza, Egypt
2Food Science Program, University of Missouri, Columbia, Missouri, USA
Received: 06 April, 2017; Accepted: 19 May, 2017; Published: 20 May, 2017
Azlin Mustapha, Food Science Program, Division of Food Systems and Bioengineering, 246 William Stringer Wing, Eckles Hall, University of Missouri, Columbia, MO 65211, USA, Tel: +1-573-882-2649; Fax: +1-573-884-7964; E-mail:
Mahmoud Ouf JM, Yuan Y, Singh P, Mustapha A (2017) Detection of Viable but Nonculturable Escherichia coli O157:H7 in Ground Beef by Propidium Monoazide real-time PCR. Int J Agric Sc Food Technol 3(2): 026-031. DOI: 10.17352/2455-815X.000018
© 2017 Mahmoud Ouf JM, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
E. coli O157; qPCR; PMA; Lactic acid; Stress
Escherichia coli O157:H7 can enter into a viable but nonculturable (VBNC) state under stress conditions. Pathogens in this dormant state may escape detection if conventional methods are employed, and potentially pose serious threats to human health. Studies have shown that many intervention and preservation processes that are commonly used in the food industry may instead induce a VBNC state rather than kill the intended pathogens. This study aimed to detect whether E. coli O157:H7, an important and dangerous foodborne pathogen, could adapt to the stress caused by lactic acid exposure by entering the VBNC state. A propidium monoazide (PMA) quantitative PCR (qPCR) method was used for detection and quantification of VBNC E. coli O157:H7 cells. The performance of this PMA-qPCR method was assessed using pure culture and ground beef samples inoculated with VBNC E. coli O157:H7 cells. The applied assay could detect as low as 103 CFU/mL VBNC E. coli O157:H7 in pure culture and 4 × 104 CFU/g VBNC cells in ground beef. Results indicate that PMA qPCR could accurately quantify E. coli O157:H7 in a VBNC state.
Introduction
Escherichia coli O157:H7 is an important foodborne pathogen that causes gastrointestinal illness as well as life-threatening diseases [1]. This pathogen can colonize the intestinal tract of cattle and make its way into beef products during slaughtering and subsequent processing. The infectious dose of E. coli O157:H7 ranges from 10-100 cells and as low as fewer than 50 viable E. coli O157:H7 cells can lead to serious outbreaks [2]. Furthermore, this pathogen has the potential to enter into the viable-but-nonculturable state [3]. In such a state, cells fail to grow and form colonies on commonly used selective media for their detection, but remain alive and retain their metabolic activities [4]. In fact, pathogenic bacteria can be avirulent in the VBNC state but regain virulence after resuscitation into culturable cells under suitable conditions [5]. Reissbrodt et al. (2002) [6], reported that VBNC cells may resuscitate in the presence of certain growth promoters or enrichments. Some VBNC cells are still virulent and even cause fatal infections, which may be due to their rapid resuscitation in suitable hosts [5,7]. In E. coli O157:H7, the expression of multiple virulence genes, including the Shiga toxin genes, stx1 and stx2 genes, can still occur in VBNC cells [8] and strains of this bacterium in the VBNC state can become culturable again in the presence of the antioxidant, oxyrase, the enterobacterial autoinducer or sodium pyruvate [9].
The distinction between viability and culturability is especially critical for pathogens, because loss of culturability may not guarantee loss of pathogenicity. If pathogenicity persists, pathogens in the dormant (VBNC) state may, in fact, pose a heretofore unrecognized public health threat [10]. The occurrence and persistence of VBNC cells that retain pathogenicity or are able to recover from this state is a public health concern since they may constitute an unrecognized source of infection [11]. In fact, because cells of E. coli O157:H7 in the VBNC state retain virulence, they should be considered as risks to public health [12].
Conventional culture-based methods involving enrichment, isolation and confirmation steps, are commonly used for detection of foodborne pathogens [13]. However, culture-based methods may considerably underestimate true bacterial cell counts when a fraction of E. coli O157:H7 cells in a sample is nonculturable [14]. Molecular methods, such as the polymerase chain reaction (PCR), are increasingly used for detection of VBNC cells [15], because they do not rely on colony growth. However, conventional PCR cannot differentiate viable cells from dead cells because DNA from dead cells can serve as a template during PCR amplification [16]. Our previous publications [17-19], have shown that DNA-intercalating agents, such as propidium monoazide (PMA) and ethidium monoazide (EMA) can be used in conjunction with a PCR assay to selectively detect viable cells. These dyes can penetrate membrane-damaged cells and covalently bind to cellular DNA, thus preventing DNA amplification from dead bacteria and enabling exclusive detection of viable cells [20-22].
This study aimed to determine if E. coli O157:H7 could adapt to the stress caused by lactic acid by entering the VBNC state, and to evaluate the applicability of a PMA-qPCR to detect and quantify VBNC cells of this pathogen. The performance of this PMA-qPCR method was assessed using pure culture and ground beef samples spiked with lactic acid induced VBNC E. coli O157:H7 cells.
Materials and Methods
Preparation of viable E. coli O157:H7 cells
E. coli O157:H7 strain 505B (a beef isolate) was obtained from the culture collection of the Food Microbiology Laboratory, University of Missouri Columbia (Columbia, MO, USA). The strain was grown overnight in Tryptic Soy broth supplemented with 0.5% yeast extract (TSBY, Difco Labs., Sparks, MD, USA) at 37 °C until the numbers reached ~109 CFU/mL as determined by plate counting on Tryptic Soy Agar (TSA) (Difco Labs.). Fresh overnight grown E. coli O157:H7 culture was serially diluted using 9 mL 0.1% peptone water, plated in TSA, and colonies enumerated after an overnight incubation at 37 oC.
PMA treatment
Serial dilutions (10-2 to 10-8) of a freshly grown culture of E. coli O157:H7 cells were prepared in 0.1% peptone water. Two milliliters of each diluted suspension were withdrawn and divided into two 1-mL suspensions in separate tubes. One set was used for DNA extraction without PMA treatment, whereas the other set was stained in the dark for 5 min with 25 μM PMA (Biotium Inc., Hayward, CA, USA), placed in ice and exposed to a 650-W halogen light at a distance of 20 cm for 10 min, as previously optimized and described by our group [17]. PMA treated cells were centrifuged at 12,000 ×g for 5 min, then washed under the same conditions in 1 mL 0.1% peptone water.
DNA isolation
DNA from the obtained cell pellets were isolated by resuspending in 100 μL of PrepMan® Ultra Sample Preparation Reagent (Applied Biosystems, Foster City, CA, USA), according to the manufacturer’s instructions with minor modifications. In order to achieve a higher DNA yield, cell suspensions were heated at 100 °C in a dry bath incubator for 20 min. Boiled cell suspensions were centrifuged at 12000 ×g for 5 min and 10 μL of the supernatant was used as the template DNA to construct standard curves for quantitative purposes by real-time PCR assay.
Real-time PCR
Primers and probes targeting E. coli O157:H7 were designed by Wang et al. [23]. pUC19 plasmid DNA was used as an internal amplification control (IAC). Primers and probes targeting pUC19 were as designed by Fricker et al., 2007 [24] (Table 1).
Real time PCR was performed in a LightCycler® 96 real-time PCR platform (Roche Diagnostics Corporation, Indianapolis, USA). A 25 μL PCR reaction mix consisted of 12.5 μL of 2× TaqMan™ Universal PCR Master Mix (Applied Biosystems), 0.5 μM of each E. coli O157:H7 primer, 0.4 μM of each IAC primer, 0.2 μM of E. coli and IAC probe, 0.25 pg of pUC19 (8.62 × 104 copies) (Promega, Madison, WI, USA), and 5 μL template DNA. Nuclease-free water (Promega) was used to adjust the reaction volume to 25 μL [17]. The real-time PCR program consisted of 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. The concentration of E. coli O157:H7 (log CFU/mL) was determined based on a standard curve (Figure 1). Data from mean values of three independent experiments with duplicates were used to calculate the coefficient of regression (R2) values.
-
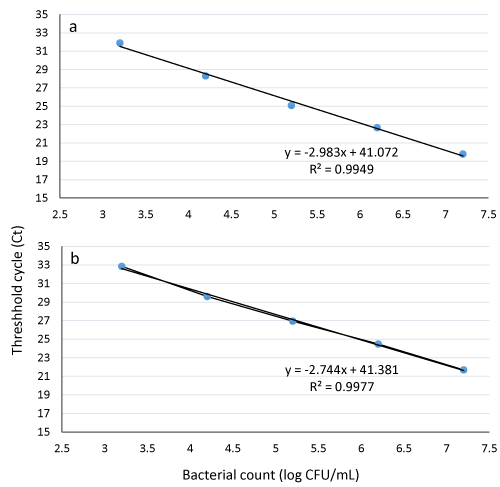
Figure 1:
Standard curves for quantifying culturable E. coli O157:H7 cells in pure culture performed with 10-fold serial dilutions using propidium monoazide (PMA) real-time PCR (a) and real-time PCR without PMA pre-treatment (b).
Induction of VBNC cells
One milliliter of freshly grown E. coli O157:H7 (ca. 109 CFU/mL) was diluted 100-fold in nutrient broth acidified with lactic acid (pH 3.0, 3.2, 3.3. 3.4 and 3.5) and subsequently incubated at 37 ºC. Samples were plated on TSA every 2 h after 10 h of incubation for up to 20 h for determining induction of the VBNC state. E. coli O157:H7 was considered to enter the VBNC state when no growth was observed on TSA plates. The same procedure was applied to obtain the counts of VBNC cells of different serial suspensions using real time PCR.
Culturability of E. coli O157:H7
Bacterial samples were serially diluted in 0.1% peptone water. One hundred microliters of appropriate dilutions were spread-plated on TSA and incubated at 37 C for 48 h to enumerate acid-stressed E. coli O157:H7 cells. When colonies ceased to occur on the agar, 1 mL of the samples was transferred to 9 mL of TSBY and incubated at 37 °C for 48 h to further test the culturability of the cells on TSA for another 48 h at 37 oC.
Preparation of artificially spiked samples
Ground beef (10% fat, 90% lean) was purchased from a local supermarket (Columbia, MO, USA) and tested for the presence of E. coli O157:H7 using standard culture methods [25]. Twenty-five grams of the ground beef samples were weighed in filtered stomacher bags (Fisherbrand, Houston, TX, USA) and artificially inoculated with different concentrations (102, 103, 104, 105, 106 and 107 CFU/g) of VBNC E. coli O157:H7. Cells were allowed to attach at room temperature for 15 min. Then, each sample was diluted with 225 mL of lactose broth (Difco Labs.), and stomached in a Stomacher 400 (Seward Lab. Systems, Inc., Bohemia, NY and USA) for 2 min at high intensity. The homogenized ground beef suspension samples were briefly centrifuged (2000 ×g for 2 min) to separate out beef and fat particles. Two milliliters of the supernatant were divided into two 1-mL portions, each of which was transferred to two fresh centrifuge tubes. Bacterial cells were collected by centrifugation at 12,500 ×g for 5 min, and the obtained cell pellets were washed in sterile distilled water under the same centrifugation conditions. Real time PCR with or without PMA pre-treatment were conducted as described in the next section.
Detection of low concentrations of VBNC E. coli O157:H7 in ground beef by PMA real-time PCR
PMA treatment of one set of samples from the previous section was applied prior to DNA extraction, using PrepMan® Ultra Sample Preparation Reagent (Applied Biosystems) as described above. One negative (un-inoculated) control for each sample was also included in the study. The extracted DNA was diluted 1:2. PCR conditions used for the two sets of DNA samples were the same as described above. Real-time PCR with IAC was performed to recognize the limit of detection of E. coli O157:H7 counts in spiked ground beef samples.
Statistical analysis
All experiments were performed in triplicate. Mean and pairwise mean comparisons were performed using Microsoft Office Excel 2007.
Results and Discussion
Quantitative differentiation of the live fraction of pathogens in raw food samples is highly critical from a public health risk perspective, as many studies have shown that under stressed conditions, major foodborne pathogens can enter a VBNC state. In this state, bacteria can remain alive for long periods of time and maintain their potential for virulence [26]. E. coli O157:H7 lost the ability to propagate after starvation in sea salt medium at 5 ºC for 70 days [3]. Wang and Doyle (1998) [27], reported that E. coli O157:H7 became nonculturable after 77 days in reservoir water or 70 days in lake water held at 25 ºC. The VBNC state of E. coli O157:H7 was also induced in 13% NaCl or after exposure to chlorine [28,29]. Previous research indicate that pH changes in media are related to a loss of culturability of bacterial cells. At 4 ºC, E. coli O157:H7 suspended in phosphate-buffered saline at pH 4 entered the VBNC state more rapidly than at pH 7 [29].
Lactic acid is widely used in food processing for reducing and eliminating pathogenic and spoilage organisms and helps to increase the shelf life of meats. It may be directly applied as a preservative ingredient or indirectly produced by natural microbial fermentation. While there have been numerous reports on the efficacy of this acid in improving food safety and quality, little is known about the occurrence of nonculturable E. coli O157:H7 in acidic foods.
In this study, the survival of E. coli O157:H7 in acidified nutrient broth was investigated in different pH and different periods in repeated experiments. Cells rapidly and significantly lost their culturability, as reflected by a rapid drop in culturable colony counts. The bacterial growth was not detected at pH 3 after 10 h, pH 3.2 after 18 h and pH 3.3 after 20 h of incubation at 37 oC (Table 2). A high degree of acid tolerance is an important feature of E. coli O157:H7 pathogen [30]. The minimum pH for E. coli O157:H7 growth is 4.0 to 4.5 [31].
-
Table 2:
Induction of VBNC E. coli O157:H7 following lactic acid treatment.
To construct the standard curves, a fresh overnight grown E. coli O157:H7 culture was serially diluted using 0.1% peptone water and culturability was detected on TSA for determination of the initial bacterial count (2 × 109 CFU/mL). Standard curves were generated (typical coefficient of determination above 98%) using PMA and non PMA real time PCR to estimate the counts of viable E. coli O157:H7. Figure 1a (PMA real-time PCR) shows a good linear reverse relationship between viable bacterial numbers and Ct values. Figure 1b (non-PMA real-time PCR) shows that the viable counts detected by culturing on TS agar are nearly in agreement with that obtained from standard curve without PMA treatment where the Ct values represent both live and dead cells (five points standard curve 103-107 cells/mL). By plugging the Ct value of an unknown sample into the standard curve generated by the PMA real time PCR (Figure 1a), the concentration of viable E. coli O157:H7 in the original sample can be estimated.
For induction of VBNC, 1 mL of an overnight E. coli O157:H7 fresh culture (2 × 109 CFU/mL) was diluted 100-fold in nutrient broth followed by lactic acid treatment for 10 hat pH 3. Data in figure 2 shows that the bacterial counts of PMA treated samples were lower than non PMA treated samples (Ct value of the PMA real-time PCR was slightly higher than that of non-PMA real-time PCR with the same numbers of cells), indicating that the PMA had removed any influence of dead cell DNA from the PCR. In non-PMA treated samples, DNA of dead cells from the lactic acid treatment was amplified during the PCR, resulting in the failure to distinguish between DNA from dead or viable cells. This confirm that PMA, a DNA intercalating dye, can only penetrate dead or membrane-compromised cells and covalently bind to cellular DNA through photolysis, rendering the DNA insoluble and inhibit PCR amplification of DNA. This agrees with Nocker et al. (2006) [21], who mentioned that PMA is less likely to permeate viable cells and probably VBNC with intact membranes, as well as with our previous study [17].
-
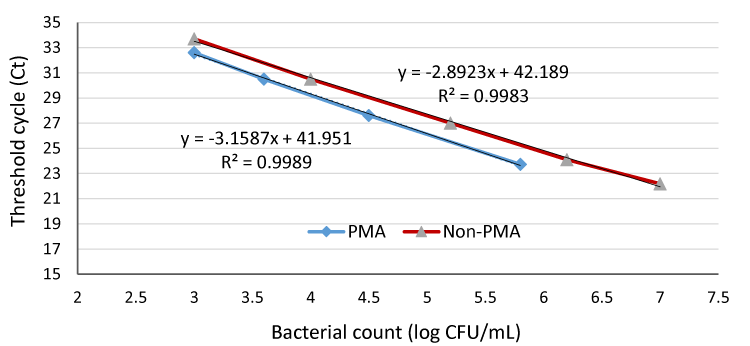
Figure 2:
Quantification of VBNC and non-viable E. coli O157:H7 in pure culture samples with PMA and non-PMA real-time PCR.
From Figure 2, we can also notice that the Ct values increased with decreasing VBNC counts (107-102 CFU/mL) in both PMA and non-PMA treated samples. The mean Ct values from replicates were not significantly different. For VBNC cells, the lowest detection limit of the PMA and non–PMA treatment was nearly 103 CFU/mL (Figure 3). A better sensitivity was reported by Xiao et al. (2013) [32] who determined that the quantification limit of a PMA–qPCR was 102 CFU/mL, providing sufficient sensitivity for detection of VBNC E. coli O157:H7 cells to no less than 100 CFU/mL.
Figure 3 shows that the quantification limits for VBNC cells were 4 × 104 and 6.3 × 103 CFU/g for PMA- and non-PMA real time PCR of spiked ground meat samples, respectively. PMA molecules could penetrate the pathogenic bacterial cells without being negatively influenced by the meat background microflora as the data showed that for all bacterial suspension examined, positive results were obtained from all trials with Ct values below 35. This would eliminate possible false-positive signals and overestimation results generated from dead cells or damaged cells that may be detected by using real time PCR alone. Nearly similar results were reported by Liu and Mustapha (2014) [17], who stated that the PMA real-time PCR could detect a range from 105 to 108 CFU/g of viable E. coli O157:H7, in ground beef samples regardless of whether dead cells were present or not. They added that substances naturally found in environmental and food samples may inhibit the amplification of target DNA in real-time PCR. This may be the reason for the relatively high detection limit of VBNC E. coli O157:H7 cells in ground beef (4 ×104 CFU/g) in our study. Španová et al. (2000) [33], reported that food samples contain many organic and inorganic substances, such as phenolic compounds, fat, enzymes, polysaccharides, proteins and salts, all of which can either inhibit PCR amplification or lead to a reduction in amplification efficiency of PCR reactions. This detection limit (4 × 104 CFU/g) in ground beef was not satisfactory with respect to the low infectious dose of E. coli O157:H7. Therefore, to use this procedure, a resuscitation or enrichment step would have to be incorporated especially when the initial concentration is low. In pure culture or spiked ground beef, the bacterial cells treated with PMA staining showed lower counts (the Ct values of PMA qPCR were 1–2 cycles higher than those of qPCR at the same numbers of cells), further highlighting the importance of an enrichment step.
-
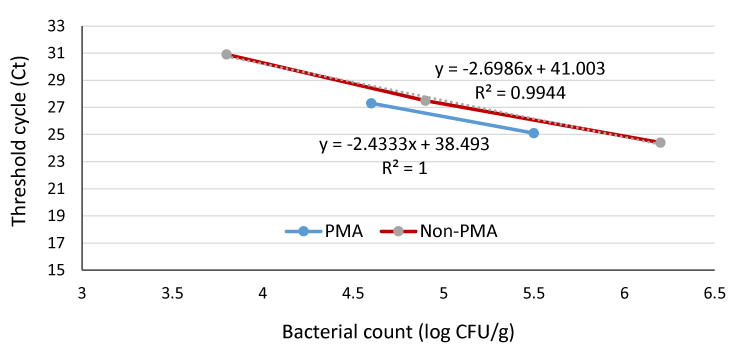
Figure 3:
Quantification of VBNC and non-viable E. coli O157:H7 in spiked ground beef samples with PMA and non-PMA real-time PCR.
The differing results between the viable count detected by PMA real-time PCR and that from the non-PMA real-time PCR (Figure 2) indicated that E. coli O157:H7 cells after lactic acid treatment entered a VBNC state. The addition of an internal amplification control (IAC) in a real-time PCR reaction system allows one to monitor the efficiency of each reaction and prevent false-negative results [34-36]). pUC19 is a small, high copy number E. coli plasmid with a molecular weight of 2686 bp. It has been previously used with success as an IAC in real-time PCR [24]. Further, a negative control (water) was added to ensure that no DNA cross-contamination occurred in the PCR reaction. A real-time PCR assay is more sensitive than traditional PCR and can be quantitative for pathogenic bacterial detection in food.
Conclusion
The results confirm that lactic acid, widely used in food processing, can induce E. coli O157:H7 to enter the VBNC state. The nonculturable cells of E. coli O157:H7 failed to propagate even in TSBY and maintained intact membranes for 10 hat pH 3 which indicate that acid-stressed E. coli O157:H7 cells exhibited signs of metabolic activity. However, a minimum of approximately 103 and 4 × 104 CFU was required for the detection of VBNC E. coli O157:H7 using PMA real-time PCR in pure culture and spiked meat samples, respectively. A resuscitative step is needed during food examination when the initial concentration of pathogens is low. We are still in need for accurate analytical method for acidified foods and more researches on resuscitation and pathogenicity of acid-induced VBNC E. coli O157:H7.
Acknowledgment
We acknowledge the US Department of Agriculture Scientific Scholars Exchange Program for funding this research.
-
-
- Carol AP (1999) The epidemiology, detection and control of Escherichia coli O157. J Sci Food Agric 79: 1367–1381. Link: https://goo.gl/CN9nF5
- Marouani-Gadri N, Firmesse O, Chassaing D, Sandris-Nielsen D, Arneborg N, et al. (2010) Potential of Escherichia coli O157:H7 to persist and form viable but non-culturable cells on a food-contact surface subjected to cycles of soiling and chemical treatment. Int J Food Microbiol 144: 96–103. Link: https://goo.gl/2Q47mY
- Rigsbee W, Simpson LM, Oliver JD (1997) Detection of the viable but nonculturable state in Escherichia coli O157:H7. J Food Safety 16: 255–262. Link: https://goo.gl/S2A9pj
- Oliver JD (2000) The public health significance of viable but nonculturable bacteria. In Nonculturable Microorganisms in the Environment 277-300. Link: https://goo.gl/GtMQrs
- Du M, Chen J, Zhang X, Li A, Li Y, et al. (2007) Retention of virulence in à viable but nonculturable Edwardsiella tarda isolate. Appl Environ Microbiol 73: 1349–1354. Link: https://goo.gl/1cyxYX
- Reissbrodt R, Rienaecker I, Romanova JM, Freestone PPE, Haigh RD, et al. (2002) Resuscitation of Salmonella enterica serovar Typhimurium and enterohemorrhagic Escherichia coli from the viable but nonculturable state by heat stable enterobacterial autoinducer. Appl Environ Microbiol 68: 4788–4794. Link: https://goo.gl/kRJfr6
- Baffone W, Citterio B, Vittoria E, Casaroli A, Campana R, et al. (2003) Retention of virulence in viable but nonculturable halophilic Vibrio spp. Int J Food Microbiol 89: 31-39. Link: https://goo.gl/LbZ3tD
- Yaron S, Matthews KR (2002) A reverse transcriptase-polymerase chain reaction assay for detection of viable Escherichia coli O157:H7: investigation of specific target genes. J Appl Microbiol 92: 633–640. Link: https://goo.gl/H1lfia
- Asakura H, Igimi S, Kawamoto K, Yamamoto S, Makino S (2005) Role of in vivo passage on the environmental adaption of enterohemorrhagic Escherichia coli O157:H7: cross-induction of the viable but nonculturable state by osmotic and oxidative stresses. FEMS Microbiol Lett 253: 243–249. Link: https://goo.gl/lj2EYQ
- Rahman I, Shahamat M, Chowdhury R (1996) Potential virulence of viable but nonculturable Shigella dysenteriae Type 1. Appl Environ Microbiol 62: 115-120. Link: https://goo.gl/zUbXY8
- Dinu LD, Bach S (2011) Induction of viable but non-culturable Escherichia coli O157:H7 in the hyllosphere of lettuce: a food safety risk factor. Appl Environ Microbiol 77: 8295-8302. Link: https://goo.gl/EsFA7f
- Oliver JD (2005) The viable but nonculturable state in bacteria. J Microbiol 43: 93–100. Link: https://goo.gl/JyzK9G
- Murakami T (2012) Filter-based pathogen enrichment technology for detection of multiple viable foodborne pathogens in one day. J Food Prot 75: 1603–1610. Link: https://goo.gl/ZkU64V
- Rowan NJ (2004) Viable but nonculturable forms of food and waterborne bacteria: QuoVadis? Trends Food Sci Technol 15: 462–467. Link: https://goo.gl/PmJBXk
- Oliver JD (2010) Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS Microbiol Rev 34: 415–425. Link: https://goo.gl/Oogkva
- Wang S, Levin RE (2006) Discrimination of viable Vibrio vulnificus cells from dead cells in real-time PCR. J Microbiol Methods 64: 1–8. Link: https://goo.gl/swlWKM
- Liu Y, Mustapha A (2014) Detection of viable Escherichia coli O157:H7 in ground beef by propidium monoazide real-time PCR. Int J Food Microbiol 170: 48–54. Link: https://goo.gl/VTUdFa
- Wang L, Li Y, Mustapha A (2009) Detection of viable Escherichia coli O157:H7 by ethidium monoazide real-time PCR. J Appl Microbiol 107: 1719-1728. Link: https://goo.gl/ssc1el
- Wang L, Mustapha A (2010) EMA-real-time PCR as a reliable method for detection of viable Salmonella in chicken and eggs. J Food Sci 75: M134-M139. Link: https://goo.gl/UCiyDm
- Lee JL, Levin RE (2009) A comparative study of the ability of EMA and PMA to distinguish viable from heat killed mixed bacterial flora from fish fillets. J Microbiol Methods 76: 93–96. Link: https://goo.gl/dCMs9m
- Nocker A, Cheung CY, Camper AK (2006) Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cell. J Microbiol Methods 67: 310–320. Link: https://goo.gl/SDPJrH
- Pan Y, Breidt F (2007) Enumeration of viable Listeria monocytogenes cells by real-time PCR with propidium monoazide and ethidium monoazide in the presence of dead cells. Appl Environ Microbiol 73: 8028–8031. Link: https://goo.gl/5H6TkI
- Wang L, Li Y, Mustapha A (2007) Rapid and simultaneous quantitation of Escherichia coli O157:H7, Salmonella and Shigella in ground beef by multiplex real-time PCR and immunomagnetic separation. J Food Prot 70: 1366-1372. Link: https://goo.gl/AhfF5F
- Fricker M, Messelha¨ußer U, Busch U, Scherer S, Ehling-Schulz M (2007) Diagnostic real-time PCR assays for the detection of emetic Bacillus cereus strains in foods and recent food-borne outbreaks. Appl Environ Microbiol 73: 1892–1898. Link: https://goo.gl/1K1Wqo
- FDA (1995) Bacteriological Analytical Manual, 8th edition. Gaitherburg, MD, USA:AOAC International
- Dinu LD, Bach S (2013) Detection of viable but non-culturable Escherichia coli O157:H7 from vegetable samples using quantitative PCR with propidium monoazide and immunological assays. Food Control 31: 268-273.Link: https://goo.gl/gMA4hq
- Wang G, Doyle MP (1998) Survival of enterohemorrhagic Escherichia coli O157:H7 in water. J Food Prot 61: 662–667. Link: https://goo.gl/2sVmdA
- Makino SI, Kii T, Asakura H, Shirahata T, Ikeda T, et al. (2000) Does enterohemorrhagic Escherichia coli enter the viable but nonculturable state in salted salmon roe? Appl Environ Microbiol 66: 5536–5539. Link: https://goo.gl/U8Y053
- Zhao L, Matthews KR (2000) Influence of starvation, temperature, and pH on culturability of enterohemorrhagic Escherichia coli O157:H7. J Food Safety 20: 193–208. Link: https://goo.gl/Hd0aj9
- Benjamin MM, Datta AR (1995) Acid tolerance of enterohemorrhagic Escherichia coli. Appl Environ Microbiol 61: 1669-1672. Link: https://goo.gl/w6kz4I
- Buchanan RL, Bagi LK (1994) Expansion of response surface models for the growth of Escherichia coli O157:H7 to include sodium nitrite as a variable. Int J Food Microbiol 23: 317-322. Link: https://goo.gl/R2wiZJ
- Xiao Xl, Tian C, Yu YG, Wu H (2013) Detection of viable but nonculturable Escherichia coli O157:H7 using propidium monoazide treatments and qPCR. Can J Microbiol 59: 157-163. Link: https://goo.gl/8OH8xn
- Španová A, Rittich B, Karpíšková R, Čechová L, Škapová D (2000) PCR identification of Salmonella cells in food and stool samples after immunomagnetic separation. Bioseparation 9: 379–384. Link: https://goo.gl/rkW6W2
- Lambertz ST, Ballagi-Pordany A, Lindqvist R (1998) A mimic as internal standard to monitor PCR analysis of food-borne pathogens. Lett Appl Microbiol 26: 9–11. Link: https://goo.gl/7fB3ct
- Hoorfar J, Cook N, Malorny B, Wagner M, De Medici D, et al. (2004) Diagnostic PCR: making internal amplification control mandatory. J Clin Microbiol 96: 221–222. Link: https://goo.gl/tGX3AC
- Murphy NM, McKauchlin J, Ohai O, Grant KA (2007) Construction and evaluation of a microbiological positive process internal control for PCR-based examination of food samples for Listeria monocytogenes and Salmonella enterica. Int J Food Microbiol 120: 110–119. Link: https://goo.gl/2EVpfJ










Table 1:
Primers, probes and PCR product sizes.
Oligonucleotide
Primer and probe sequence (5′–3′)
Conc.
Product Size
Reference
E. coli O157:H7 primer 1
5′-TTGACCCACACTTTGCCGTAA-3′
0.5 μM
226 bp
Wang et al., 2007
E. coli O157:H7 primer 2
5′-GCGAAAACTGTGGAATTGGG-3′
0.5 μM
Wang et al., 2007
E. coli O157:H7 probe
5′-5HEX-TGACCGCATCGAAACGCAGCT-BHQ1-3′
0.2 μM
Wang et al., 2007
IAC_for
5′-GCAGCCACTGGTAACAGGAT-3′
0.4 μM
118 bp
Fricker et al. 2007
IAC_rev
5′-GCAGAGCGCAGATACCAAAT-3′
0.4 μM
Fricker et al. 2007
IAC_probe
5′-56FAM-AGAGCGAGGTATGTAGGCGG-BHQ1-3′
0.2 μM
Fricker et al. 2007