Author(s):
JF Correia-Silva1, RG Resende1, MHNG Abreu2, AL Teixeira3, MM Teixeira4, H Bittencourt5, RS Gomez1 and TA Silva1*
1Department of Oral Surgery and Pathology, School of Dentistry, Federal University of Minas Gerais, Belo Horizonte, Minas Gerais, Brazil
2Department of Community and Preventive Dentistry, School of Dentistry, Universidade Federal de Minas Gerais, Belo Horizonte, Brazil
3Department of Clinical Medicine, Universidade Federal de Minas Gerais, Faculty of Medicine Belo Horizonte, Minas Gerais, Brazil
4Department of Biochemistry and Immunology, Institute of Biological Sciences, Universidade Federal de Minas Gerais, Belo Horizonte, Minas Gerais, Brazil
5Hematopoietic Stem Cell Transplantation Unit, Department of Hematology, Hospital das Clínicas, Universidade Federal de Minas Gerais, Belo Horizonte, Minas Gerais, Brazil
Received: 11 April, 2016; Accepted: 15 June, 2016; Published: 17 June, 2016
Prof. Tarcilia Aparecida Silva, Departamento de Clínica Patologia e Cirurgia, Faculdade de Odontologia, Universidade Federal de Minas Gerais, Av. Antônio Carlos 6627, Pampulha, Belo Horizonte, Minas Gerais 31270-901, Brazil, Tel: +55 31 3409 2478; Fax: +55 31 34092430; E-mail:
Correia-Silva JF, Resende RG, Abreu MHNG, Teixeira AL, Teixeira MM, et al. (2016) Cytokine Production and Human Cytomegalovirus Load in Allogeneic Hematopoietic Stem Cell Transplantation Outcome. Int J Immunother Cancer Res 2(1): 003-010. 10.17352/2455-8591.000008
© 2016 Correia-Silva JF et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Allogeneic HSCT; Cytokines; HCMV; Survival
allo-HSCT: allogeneic Hematopoietic Stem Cell Transplantation; GVHD: Graft-Versus-Host Disease; HCMV: Human Cytomegalovirus; IL: Interleukin; IFN: Interferon; TNF-α: Tumor Necrosis Factor-α; HLA: Human Leukocyte Antigen; HR: Hazard Ratio;
Objective: To investigate the impact of cytokine levels (IL-1β, IL-6, IL-10, IFN-γ and TNF-α) and Human cytomegalovirus (HCMV) load in the saliva and blood on the survival of allogeneic hematopoietic stem cell transplantation (allo-HSCT) recipients.
Study Design: Samples were obtained from 63 patients 7 days before and 21 days after allo-HSCT. Cytokine levels were assessed by ELISA, and HCMV load was determined by real-time PCR.
Results: The increase of IL-6 in the saliva and the reduction of IFN-γ in the blood before allo-HSCT were associated with increased risk of death. Moreover, the increase of IL-6 in the blood and of HCMV in the saliva after allo-HSCT were also associated with increased risk of death.
Conclusions: Cytokine levels and HCMV load were associated with the increased risk of death. The findings suggest a potential function of these biomarkers in the determination of allo-HSCT survival.
Introduction
Allogeneic hematopoietic stem cell transplantation (allo-HSCT) is performed for a range of disorders, including hematological malignancy, severe aplastic anemia and genetic diseases. The attractive aspect of this therapeutic strategy is the development of potent donor T-cell-mediated immune responses that can eliminate malignant cells, which has traditionally been termed the graft versus leukemia effect. Although allo-HSCT provides the only curative therapy for many patients with malignant and non-malignant diseases, it is also associated with a high incidence of treatment-related morbidity and mortality [1,2].
Allo-HSCT recipients generally present three disturbance levels in the first weeks after allo-HSCT. These disturbances potentially can stimulate a massive release of inflammatory cytokines [3]. First, baseline circulating inflammatory cytokines, such as tumor necrosis factor-α (TNF-α) and interleukin-1 (IL-1), could be increased as a result of disease, therapy, comorbidities or ongoing infections that occur before allo-HSCT. Second, immediately after allo-HSCT, the immune dysfunction caused by the conditioning regimen, which may produce tissue injury and consequently the production of inflammatory cytokines, may affect the reconstitution of white blood cells. Third, after allo-HSCT, activated, mature donor T-cells can set off a cytokine response that in turn stimulates host T-cell responses related to the onset of acute graft-versus-host disease (aGVHD) [3,4]. Immune dysfunction following allo-HSCT involves activated T-cells secreting interferon (IFN)-γ as well as other cytokines, which activate monocytes and dendritic cells. This process appears to be associated with the induction of aGVHD, resulting in significant morbidity and mortality. In addition, a prolonged immune deficiency characterized by lymphopenia and susceptibility to infection can occur [2]. Deregulation of inflammatory cytokines is also associated with multiple symptoms (pain, sleep disturbance and depression) during neutropenia after allo-HSCT [3]. Interleukin-6 (IL-6) is an important pro-inflammatory cytokine involved in immune-mediated complications after allo-HSCT [5].
Human cytomegalovirus (HCMV) infection remains one of the most important causes of morbidity and mortality among allo-HSCT recipients [6,7]. Infection by HCMV may result not only in HCMV disease, but is also associated with other adverse effects due to its immunomodulatory actions; this might result in increased risk of bacterial, fungal and viral infections, and also of aGVHD [8].
Allo-HSCT outcomes could be affected by factors such as HCMV infection, recipient/donor histocompatibility, patient/donor gender, recipient age, GVHD and cytokine production [9,10]. The purpose of this study was to investigate the impact of cytokine levels (IL-1β, IL-6, IL-10, IFN-γ and TNF-α) and HCMV load in the saliva and blood, before and after allo-HSCT, on recipient survival.
Materials and Methods
The study protocol was approved by the Research Ethics Committee of the institution (Process # ETIC 097/06). Informed consent was obtained from all the patients or from parents, if the patient was less than 18 years.
Patients and samples
Sixty-three consecutive allo-HSCT patients from Hospital das Clínicas of Universidade Federal de Minas Gerais (HC-UFMG), were included in this prospective study. The patients were prepared for allo-HSCT according to the protocols of the Stem Cell Transplant Unit at HC-UFMG, which vary according to type and status of the disease. Cyclosporine, in combination with either methotrexate or mycophenolate mofetil, was used for GVHD prophylaxis. Methylprednisolone in combination with Cyclosporine was used for GVHD treatment. Antigenemia assay was used routinely for preemptive ganciclovir treatment of HCMV infection when 2 or more neutrophils’ positive nuclei for the HCMV lower matrix phosphoprotein pp65 in a cytospin preparation of 105 blood leukocytes after neutrophil recovery (‡500 neutrophils mm3) were detected, according to the protocols of the allo-HSCT Unit of HC-UFMG. This study was approved by the Institutional Ethics Committee for human subjects.
Demographic information, as well as clinical and laboratory data, were available from the database of the HC-UFMG. This clinical information includes the underlying disease, the source of the stem cells, the gender and age of the patient, the gender of the donor, the HCMV serostatus of the donor and recipient before transplantation, conditioning regimes, Human leukocyte antigen (HLA) matching and pp65 antigenemia status.
Saliva and blood samples from 63 recipients, were taken in two different stages [11], seven days before (stage 2) and 21 days after (stage 3) HSCT. Patients were followed for one year after allo-HSCT, or until the death of the recipient.
Enzyme-Linked Immunosorbent Assay (ELISA)
Saliva and blood from 63 patients who submitted to allo-HSCT were collected for quantification of cytokines by ELISA. Oral fluid was collected using Salivette® neutral cotton swabs (Sarstedt, Nümbrecht, Germany). Recovery of the saliva was achieved by centrifuging the container at 1500 rpm for 10 minutes. Saliva samples were diluted (1:1) in PBS (0.4 mM NaCl and 10 mM NaPO4) containing protease inhibitors (0.1 mM PMSF, 0.1 mM benzethonium chloride, 10 mM EDTA and 0.01 mg/mL aprotinin A) and 0.05% Tween-20, and frozen at -20°C until analysis. Four milliliters of peripheral blood were collected in anticoagulant-free tubes and centrifuged at room temperature for 30 min. The recovered serum was stored in small aliquots at -20°C until the cytokines were quantified. Concentrations of IL-1β, IL-6, IL-10, IFN-γ and TNF-α in the saliva and blood were determined using commercially available, quantitative sandwich ELISA kits (DuoSet, R&D Systems, Minneapolis, MN, USA), according to the manufacturers’ instructions. The detection ranges were 3000-46 pg/ml for IL-1β, 600-9 pg/ml for IL-6, 4000-62 pg/ml for IL-10, 1000-15 pg/ml for IFN-γ, and 1000-15 pg/ml for TNF-α. Values below the detection limits were assumed to be zero. Concentrations were expressed as pg/mL for blood. The total protein in the saliva samples was measured using the Bradford method, according to the BSA standard (Fermentas Life Sciences, Vilnius, Lithuania), and concentrations were expressed as mg/ml. Total protein concentration was used to correct the saliva cytokine values for each sample. The saliva sample values, corrected by the total protein values, were expressed in pg/mg protein.
qPCR
Saliva and blood from the first 30 patients enrolled in this study were collected for real-time PCR assays. Oral fluid was collected by swab, as previously described [7]. Four milliliters of peripheral blood were collected in an EDTA tube for the PCR assay and stored at -20°C until processing. Total genomic DNA was extracted from saliva and whole-blood samples using a QIAamp DNA Blood Mini Kit (Qiagen, Valencia, CA, USA) and stored at -20°C until used. Two hundred nanograms of extracted DNA were used for HCMV real-time PCR. The HCMV-specific PCR primers used in the assay were selected from the US17 region of HCMV AD169 [12]. Quantification of HCMV DNA was performed using SYBR Green PCR Core Reagents (PE Applied Biosystems, Foster City, CA, USA). PCR was performed with an ABI Prism 7900 instrument (PE Applied Biosystems, Foster City, CA, USA), using 96-well plates. The mean value of the duplicates was used in calculations of HCMV DNA.
Statistical analysis
Overall survival was calculated using the Kaplan-Meier method. Time to death after allo-HSCT was determined initially compared using the log-rank test for the following variables: age of patient, recipient gender, donor gender, recipient/donor gender, primary disease, stem cell source, HLA matching, clinical systemic aGVHD, pp65 antigenemia, conditioning regimen, IL-1β, IL-6, IL-10, IFN-γ and TNF-α levels before and after allo-HSCT, and HCMV load in saliva and blood before and after allo-HSCT. Based on p values lower than 0.25 the variables were included in the Cox proportional hazards models [13,14]. Four models were created. Model 1 presented the following variables: recipient/donor gender, stem cell source, clinical systemic aGVHD, and IL-6 levels in saliva before allo-HSCT. Model 2 presented the following variables: recipient/donor gender, stem cell source, and clinical systemic aGVHD. Model 3 presented the following variables: recipient/donor gender, stem cell source, clinical systemic aGVHD, and IFN-γ levels in blood before allo-HSCT. Model 4 presented the following variables: recipient/donor gender, stem cell source, clinical systemic aGVHD, and IL-6 levels in blood after allo-HSCT. Survival time after HSCT was calculated until the last follow-up or until death. The continuous variable was categorized for generating a new categorical variable with group numbers based on the median of this variable. The Kaplan-Meier curves were generated with the new categorical variables. Statistical analyses were performed using SPSS (SPSS Inc., version 16.0, Chicago, IL), and p values ≤ 0.05 were considered statistically significant.
Results
Demographic data
Allo-HSCT recipients were followed from 7 days until one year after allo-HSCT, or until the death of the recipient. Of the 63 allo-HSCT recipients, 38 (60%) remained alive one year after HSCT. The survival median of the 25 deceased patients after allo-HSCT was 68 days (range 17-240). Patient and transplant characteristics, conditioning therapy, aGVHD, and survival univariate analyses are shown in Table 1.
The twenty five patients who died include the 3 patients that presented relapse. Among the transplantation-related causes: six presented GVHD, five presented bacterial infection, three presented viral infection, five presented fungal infection, two presented acute respiratory distress syndrome, and one presented acute liver insufficiency. Three recipients developed HCMV disease: two developed pneumonia and one developed enterocolitis.
The average age of the patients studied was 30 years (range 5-56). Of the 63 patients, 35 (55%) were men. Compared to female recipients, male recipients demonstrated an increased risk of death after allo-HSCT (hazard ratio (HR): 4.271; 95% CI = 1.599-11.407, p= 0.004) (Figure 1A). Transplantation for a male donor to a female recipient was associated with a reduced risk of death after allo-HSCT (HR: 0.6650; 95% CI = 0.4689-0.9431, p= 0.022) (Figure 1B). We also observed a higher incidence of death after HSCT among patients receiving peripheral blood stem cells (PBSC), a source of hematopoietic stem cells for transplant (17/26; 65%) (HR: 3.8424; 95% CI = 1.6527-8.9328, p=0.002) (Figure 1C); also, among patients developing systemic aGVHD (HR: 2.7504; 95% CI = 1.2079-6.2627, p=0.0160) (Figure 1D).
-
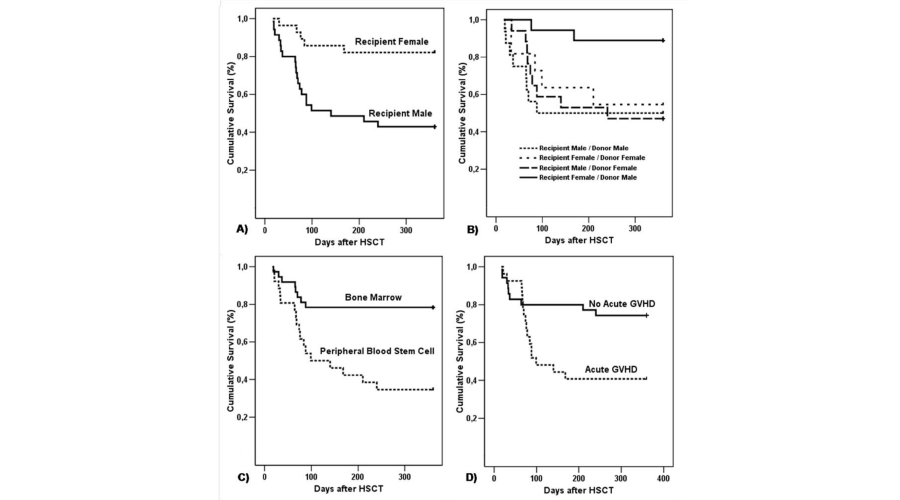
Figure 1:
Kaplan-Meier estimate of overall survival after allo-HSCT. A) Recipient gender. B) Recipient/ Donor gender. C) Stem cell source. D) Acute GVHD.
Cytokine levels
IL-1β, IL-6, IL-10, IFN-γ and TNF-α levels in the saliva and blood of the 63 allo-HSCT recipients were determined before and after allo-HSCT. The influence of cytokine levels before (7 days) and after (21 days) allo-HSCT on the patients’ survival was investigated and is shown in Table 2.
-
Table 2:
Survival according to cytokine levels in the saliva and blood before and after HSCT– Univariate analysis:
Deceased allo-HSCT recipients presented higher IL-6 levels in their saliva before allo-HSCT, compared with surviving patients (HR = 1.0028; 95% CI = 1.0001-1.0055, p= 0.045) (Table 2). Blood IL-6 levels before transplantation were also higher among patients who died, compared to patients who survived, but this difference was not statistically significant. Significantly increased levels of IL-6 in the blood after allo-HSCT were observed in recipients who died, relative to those who survived (HR = 1.0005; 95% CI = 1.0001-1.0009, p= 0.003) (Table 2).
Reduced IFN-γ levels in the blood before allo-HSCT also affected patient survival (HR = 1.0003; 95% CI = 1.0001-1.0005, p= 0.015). The median level of IFN-γ in deceased recipients was lower than in living recipients (Table 2).
Time to death after allo-HSCT was calculated using the Kaplan–Meier method for the following: IL-6 level in saliva seven days before allo-HSCT (Low level/below median - ≤ 8 pg/mg protein; High level/above median - > 8.1 pg/mg protein) (Figure 2A); IL-6 level in blood 21 days after allo-HSCT (Low level/below median - ≤ 41 pg/ml; High level/above median - > 41.1 pg/ml) (Figure 2B); and, IFN-≤ level in blood seven days before allo-HSCT (Low level/below median - = 0 pg/ml; High level/above median - > 0 pg/ml) (Figure 2C).
-
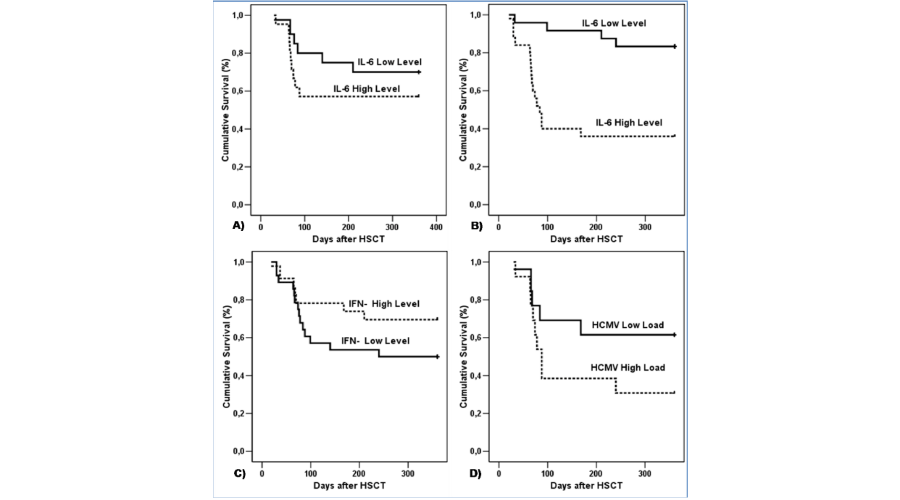
Figure 2:
Kaplan-Meier estimate of overall survival after allo-HSCT. A) IL-6 level in saliva 7 days before allo-HSCT (Low level/below median - 8 pg/mg protein; High level/above median - > 8 pg/mg protein). B) IL-6 level in blood 21 days after allo-HSCT (Low level/below median - 41pg/ml; High level/above median - >41 pg/ml). C) IFN- level in blood 7 days before allo-HSCT (Low level/below median - = 0 pg/ml; High level/above median - >0 pg/ml). D) HCMV load in saliva 21 days after allo-HSCT (Low level/below median 0.7 copies/200ng DNA; High level/above median ≥ 0.7 copies/200ng DNA).
Human cytomegalovirus (HCMV)
The results of the analysis of the impact of the HCMV load detected by real-time PCR in the saliva and blood, on survival after allo-HSCT, are shown in Table 3.
Univariate analysis showed that deceased recipients had a higher HCMV load in their saliva after HSCT (HCMV load median, 1.9 copies/200 ng DNA; range, 0-902), relative to living recipients (HCMV load median, 0 copy/200 ng DNA; range, 0-32) (HR = 1.0037; 95% CI = 1.0004-1.0066, p= 0.023) (Table 3). Time to death after allo-HSCT was also calculated using the Kaplan–Meier method for the HCMV load in saliva after HSCT (HCMV load in saliva 21 days after allo-HSCT (Low level/below median - ≤ 0.7 copies/200ng DNA; High level/above median ≥ 0.71 copies/200ng DNA) (Figure 2D).
-

Table 3:
Survival according to HCMV load by real-time PCR – Univariate analysis
Multivariate analysis
The multivariate analysis demonstrated a significant association among stem cell source, IL-6 levels in the saliva before allo-HSCT, blood IFN-γ levels before allo-HSCT, blood IL-6 levels after allo-HSCT, and patient survival. There is an increased risk of death for patients receiving a PBSC transplantation (HR: 6.1174; 95% CI 1.9257–19.4327; p=0.002); for patients with higher IL-6 levels in their saliva before allo-HSCT (HR: 1.0027; 95% CI 1.0001-1.0053; p=0.035); for patients with low IFN-γ levels in their blood before allo-HSCT (HR: 1.0003; 95% CI 1.0003-1.0005; p=0.015); and for patients with high IL-6 levels in their blood 21 days after allo-HSCT (HR: 1.0003; 95% CI 1.0001-1.0007; p=0.024) (Table 4).
-

Table 4:
Survival according to adjusted model of Cox proportional hazards multivariate models.
Discussion
In the present study we evaluated the impact of recipient gender, patient/donor gender, stem cell source, aGVHD and cytokine levels on allo-HSCT survival. Previous studies have shown that recipient and/or donor gender can affect recipient survival [15, 16]. Male patients demonstrated an increased risk of death after allo-HSCT relative to female patients. In addition, a male donor with a female recipient was associated with reduced risk of death after allo-HSCT. One possible explanation is the microchimerism, which consists on the presence of circulating donor and recipient hematopoietic cells. Studies showed that the male donor cells may initiate GVHD [15,17]. In agreement with the European Group for Blood and Marrow Transplantation, male recipients with female donors presented the worst outcome after HSCT, relative to all other donor-recipient gender combinations [12]. In another study, female donors were associated with an increased risk of patient death [18].
We found that the survival rates of allo-HSCT recipients were also influenced by the source of hematopoietic stem cells; and, it was higher for BM, in comparison to PBSC, transplantation. We also observed that the presence of aGVHD was associated with an increased risk of death after allo-HSCT. PBSC transplantation is associated with faster blood cell and immunity recoveries. However, PBSC transplantation has also been found to be associated with increased aGVHD incidence and severity [9]. The mobilization of the peripheral blood grafts contain more T-cells than marrow suggesting a potential for development of the GVHD [19]. GVHD is a complication classically associated with morbidity and mortality after HSCT [20].
Cytokines are an important part of saliva, and regulate oral cavity homeostasis. Altered salivary levels of cytokines have been found in patients with various diseases, and may reflect oral and/or systemic conditions [21]. Deceased allo-HSCT recipients presented higher IL-6 levels in saliva before allo-HSCT, in comparison with surviving patients. The multivariate analysis also demonstrated an increased risk of death for patients with high IL-6 levels in their saliva before allo-HSCT. Salivary IL-6 is an important inflammatory mediator involved in the response to localized mucosal inflammation; high levels of IL-6 have been shown to be positively associated with oral chronic GVHD severity, ulceration and erythema [22]. Increased IL-6 levels could imply an increased risk for developing oral complications, such as xerostomia and mucositis, which cause significant morbidity and potential mortality for patients undergoing allo-HSCT [23]. Thus, we can consider that the salivary IL-6 levels before allo-HSCT may reflect a local condition resulted from conditioning regimen and may serve as a useful monitor of oral mucosal damage. Further studies are necessary to investigate the function of salivary IL-6 as a potential biomarker of allo-HSCT survival.
We observed increased levels of IL-6 in the blood after allo-HSCT among patients who died, in comparison with patients who survived. However, we did not observed significant impact of blood IL-6 levels before allo-HSCT in survival recipient´s. This result suggests that IL-6 levels in the blood after allo-HSCT have a significant impact on the survival of patients. In accordance with previous reports, recipients with functional, single-nucleotide polymorphisms in the promoter region of the IL-6 gene produced high serum levels of IL-6 and presented decreased survival rates after allo-HSCT [5]. IL-6 is a pleiotropic cytokine implicated in both innate and acquired immune responses, and is an important pro-inflammatory mediator in immune-mediated complications of allo-HSCT [5,24]. IL-6 also plays multiple roles during the subsequent development of acquired immunity against incoming pathogens, including regulation of cytokine and chemokine gene expression [24]. Previous results suggested that IL-6 seemed to be the most important cytokine during the initial phase of the inflammatory process, as it was the earliest cytokine to increase in expression in the first week after HSCT [25]. In response to conditioning and stem cell infusion, serum levels of IL-6 increased during the initial 30 days after allo-HSCT (the acute phase), and recipients with early complications presented higher IL-6 levels during the first week after transplantation [3,25]. The inflammatory effects of IL-6 have also been implicated in the failure of organs in allo-HSCT recipients. Moreover, an overproduction of IL-6 during a course of GVHD has also been observed [26].
Reduced IFN-γ levels in the blood before allo-HSCT also affected patient survival. IFN-γ is a potent proinflammatory cytokine produced by CD4 T-cell subset 1 (Th1) cells, cytolytic CD8 effector T-cells and NK cells. It plays important and complex roles in both innate and adaptive immune responses [27,28]. A previous study suggested that IFN- plays a role in T-cell homeostasis by regulating T-cell expansion and survival, and activation-induced cell death [29]. IFN-γ also acts in anti-microbial response, antigen processing, inflammation, growth suppression, cell death, tumor immunity and autoimmunity [28]. Thus, a deficiency in IFN-γ production may increase the hosts’ susceptibility to infections and tumors [27,28]. Reduced IFN-γ levels could be deleterious for patients after allo-HSCT, as this cytokine may facilitate graft-versus-leukemic effects and even inhibit GVHD by donor CD4 T-cells [27]. Another study described that three of 48 patients developed HCMV disease, and none of them had detectable INF-γ production [30]. For these reasons, reduced IFN-γ levels in the blood before allo-HSCT correlated negatively with recipient survival.
Patients with a high HCMV load in the saliva after allo-HSCT presented an increased risk of death. HCMV is commonly detected in the salivary glands, endothelial cells and leucocytes, but not in oral mucosa epithelial cells [31]. HCMV presence in the oral cavity probably represents the production of an infectious virus by the salivary glands, and its subsequent shedding in the saliva. HCMV may be detectable in blood samples of patients with asymptomatic infection, who never progress to disease, because of the high sensitivity of real time PCR [32]. In addition, as PCR of blood requires 247–1650 copies of HCMV DNA per ml and 500 DNA copies per 100 ng of total leucocytes, DNA is equivalent to 10,000 cells [33], the identification of HCMV before engraftment (usually before day +30) limits early HCMV diagnosis. Thus, the identification of HCMV in the saliva could be advantageous to the diagnosis of reactivation of this virus in the period before engraftment. Previous studies have shown that high HCMV DNA in whole blood is associated with the development of HCMV infection [34]. HCMV is an important factor for morbidity and long-term outcome after allo-HSCT [35]. It has also been reported that immunosuppression related to allo-HSCT affects HCMV reactivation in the oral cavity [6,7]. In our first study [6], we did not observe any impact of HCMV presence in the saliva on recipient survival, after allo- HSCT. In this present study, however, we showed the impact of the HCMV load in saliva on patient survival. The differences might result from the study design. In addition, while in the first study we used conventional PCR [6], in the present study we used real time PCR. Furthermore, studies suggest that GVHD and HCMV replication are pathogenetically associated. HCMV may influence the development of GVHD, and GVHD and its treatment may put patients at risk for HCMV replication [36]. HCMV infection affects allograft outcomes by affecting cellular and humoral responses, including plasma cytokine production [37]. HCMV replication in renal transplant recipients is associated with increased IL-6 and decreased IFN-γ in mononuclear cell levels in plasma and peripheral blood [37,38]. HCMV DNA levels, detected by real-time PCR in whole blood, represent a precise method for determining viral load kinetics, which is useful for predicting HCMV disease [34]. A previous study showed that the HCMV load in the saliva, detected by real-time PCR, had similar kinetics as compared with blood. Moreover, this study also indicated that saliva can be a useful source for real-time PCR analysis, for the monitoring and diagnosis of HCMV disease, among patients submitted to HSCT [7].
In our previous study, a positive correlation between HCMV DNA loads in saliva and blood was achieved in patients submitted to allo-HSCT [7]. However, in patients with Sjögren’s syndrome, no association between serum and salivary levels of cytokines was observed [39,40]. This is the first study that assesses, concomitantly, the impact of saliva and blood cytokine levels on survival of allo-HSCT recipients. The results might be influenced by the relative small number of HSCT patients. Thus the clinical significance of these markers needs to be further assessed in larger populations.
In summary, cytokine levels and HCMV load were associated with the increased risk of death. These findings suggest a potential function for these biomarkers in the determination of allo-HSCT survival. Further studies are necessary to investigate the function of salivary IL-6 as a potential biomarker for allo-HSCT survival and its possible association with salivary HCMV load.
Acknowledgment
This study was supported by grants from the Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG) and from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Brazil. TA Silva, MM Teixeira, AL Teixeira and RS Gomez are research fellows of CNPq.
-
-
- Martin PJ, Counts GW Jr, Appelbaum FR, Lee SJ, Sanders JE, et al. (2010) Life expectancy in patients surviving more than 5 years after hematopoietic cell transplantation. J Clin Oncol 28: 1011-1016.
- Talmadge JE (2003) Hematopoietic stem cell graft manipulation as a mechanism of immunotherapy. Int Immunopharmacol 3: 1121-1143.
- Wang ZY, Chen Z (2008) Acute promyelocytic leukemia: from highly fatal to highly curable. Blood 2008:111:2505-2515.
- Basara N, Kiehl MG, Fauser AA (2001) New therapeutic modalities in the treatment of graft-versus-host disease. Crit Rev Oncol Hematol 38: 129-138.
- Ambruzova Z, Mrazek F, Raida L, Faber E, Onderkova J, et al. (2008) Association of IL-6 gene polymorphism with the outcome of allogeneic haematopoietic stem cell transplantation in Czech patients. Int J Immunogenet 35: 401-403.
- Correia-Silva JF, Victória JM, Guimarães AL, Salomão UE, de Abreu MH, et al. (2007) Cytomegalovirus shedding in the oral cavity of allogeneic haematopoietic stem cell transplant patients. Oral Dis 13: 163-169.
- Correia-Silva JF, Bruna-Romero O, Resende RG, Miranda LPM, Oliveira FE, et al. (2010) Saliva as a source of HCMV DNA in allogeneic stem cell transplantation patients. Oral Dis 16: 210-216.
- Behrendt CE, Rosenthal J, Bolotin E, Nakamura R, Zaia J, et al. (2009) Donor and recipient CMV serostatus and outcome of pediatric allogeneic HSCT for acute leukemia in the era of CMV-preemptive therapy. Biol Blood Marrow Transplant 15: 54-60.
- Bittencourt H, Lopes M, de Macedo AV, Teixeira ER, Gomes GG, et al. (2009) A retrospective comparison of allogeneic peripheral blood stem cell versus bone marrow transplantation. Hematol Oncol Stem Cell Ther 2: 272-277.
- Terwey TH, Hemmati PG, Martus P, Dietz E, Vuong LG, et al. (2010) A modified EBMT risk score and the hematopoietic cell transplantation-specific comorbidity index for pre-transplant risk assessment in adult acute lymphoblastic leukemia. Haematologica 95: 810-818.
- Majorana A, Schubert MM, Porta F, Ugazio AG, Sapelli PL (2000) Oral complications of pediatric hematopoietic cell transplantation: diagnosis and management. Support Care Cancer 8: 353-365.
- Machida U, Kami M, Fukui T, Kazuyama Y, Kinoshita M, et al. (2000) Real-time automated PCR for early diagnosis and monitoring of cytomegalovirus infection after bone marrow transplantation. J. Clin. Microbiol 38: 2536-2542.
- Hosmer DW, Lemesshow S (2000) Supplied logistic regression. 2nd ed. New York: Wiley-Inter science Publication.
- Kalbfleisch JD, Prentice RL (2002) The statistical analysis of failure time data. 2nd ed. New York: John Wiley & Sons.
- Souza LN, Carneiro MA, de Azevedo WM, Gomez RS (2004) Vascular endothelial growth factor (VEGF) and chronic graft-versus-host disease (cGVHD) in salivary glands of bone marrow transplant (BMT) recipients. J Oral Pathol Med 33: 13-16.
- Gratwohl A, Stern M, Brand R, Apperley J, Baldomero H, et al. (2009) Risk score for outcome after allogeneic hematopoietic stem cell transplantation: a retrospective analysis. Cancer 115: 4715-4726.
- Kuroki M, Okayama A, Nakamura S, Sasaki T, Murai K, et al. (2002) Detection of maternal-fetal microchimerism in the inflammatory lesions of patients with Sjögren's syndrome. Ann Rheum Dis 61: 1041-1046.
- De Souza CA, Vigorito AC, Ruiz MA, Nucci M, Dulley FL, et al. (2005) Validation of the EBMT risk score in chronic myeloid leukemia in Brazil and allogeneic transplant outcome. Haematologica 90: 232-237.
- Lickliter JD, McGlave PB, DeFor TE, Miller JS, Ramsay NK, et al, (2000) Matched-pair analysis of peripheral blood stem cells compared to marrow for allogeneic transplantation. Bone Marrow Transplant 26: 723-728.
- Mielcarek M, Storer BE, Boeckh M, Carpenter PA, McDonald GB, et al. (2009) Initial therapy of acute graft-versus-host disease with low-dose prednisone does not compromise patient outcomes. Blood 113: 2888-2894.
- Boras VV, Lukac J, Brailo V, Picek P, Kordić D, et al. (2006) Salivary interleukin-6 and tumor necrosis factor-alpha in patients with recurrent aphthous ulceration. J Oral Pathol Med 35: 241-243.
- Fall-Dickson JM, Mitchell SA, Marden S, Ramsay ES, Guadagnini JP, et al. (2010) Oral Symptom Intensity, Health-Related Quality of Life, and Correlative Salivary Cytokines in Adult Survivors of Hematopoietic Stem Cell Transplantation with Oral Chronic Graft-Versus-Host Disease. Biol Blood Marrow Transplant 16: 948-956.
- Bogusławska-Kapała A, Cackowska-Lass A, Balon J, Hellmann A, Kochańska B (2006) Saliva secretion and abnormal moistening of oral mucosa after bone marrow transplantation. Bull Group Int Rech Sci Stomatol Odontol 47: 1-5.
- Zedtwitz-Liebenstein K, Jaksch P, Burgmann H, Friehs H, Hofbauer R, et al. (2009) Evaluation of interleukin-6 and interleukin-10 in lung transplant patients with human cytomegalovirus infection. Clin Transplant 23: 687-691.
- Min CK, Lee WY, Min DJ, Lee DG, Kim YJ, et al. (2010) Correlations of HHV-6 viral load and plasma IL-6 concentration with HHV-6 encephalitis in allogeneic stem cell transplant recipients. Bone Marrow Transplant 45: 129-136.
- Chen X, Das R, Komorowski R, Beres A, Hessner MJ, et al. (2009) Blockade of interleukin-6 signaling augments regulatory T-cell reconstitution and attenuates the severity of graft-versus-host disease. Blood 114: 891-900.
- Yang YG, Wang H, Asavaroengchai W, Dey BR (2005) Role of Interferon-gamma in GVHD and GVL. Cell Mol Immunol 2: 323-329.
- Saha B, Jyothi Prasanna S, Chandrasekar B, Nandi D (2010) Gene modulation and immunoregulatory roles of interferon gamma. Cytokine 50: 1-14.
- Refaeli Y, Van Parijs L, Alexander SI, Abbas AK (2002) Interferon gamma is required for activation-induced death of T lymphocytes. J Exp Med 196: 999-1005.
- Avetisyan G, Larsson K, Aschan J, Nilsson C, Hassan M, et al. (2006) Impact on the cytomegalovirus (CMV) viral load by CMV-specific T-cell immunity in recipients of allogeneic stem cell transplantation. Bone Marrow Transplant 38: 687-692.
- Kano Y, Shiohara T (2000) Current understanding of cytomegalovirus infection in immunocompetent individuals. J Dermatol Sci 22: 196-204.
- Tanaka N, Kimura H, Iida K, Saito Y, Tsuge I, et al. (2000) Quantitative analysis of cytomegalovirus load using a real-time PCR assay. J Med Virol 60: 455-462.
- Boriskin YS, Fuller K, Powles RL, Vipond IB, Rice PS, et al. (2002) Early detection of cytomegalovirus (CMV) infection in bone marrow transplant patients by reverse transcription-PCR for CMV spliced late gene UL21.5: a two site evaluation. J Clin Virol 24: 13-23.
- Allice T, Cerutti F, Pittaluga F, Varetto S, Franchello A, et al. (2008) Evaluation of a novel real-time PCR system for cytomegalovirus DNA quantitation on whole blood and correlation with pp65-antigen test in guiding pre-emptive antiviral treatment. J Virol Methods 148: 9-16.
- Gerna G, Lilleri D, Caldera D, Furione M, Zenone Bragotti L, et al. (2008) Validation of a DNAemia cutoff for preemptive therapy of cytomegalovirus infection in adult hematopoietic stem cell transplant recipients. Bone Marrow Transplant 41: 873-879.
- Cantoni N, Hirsch HH, Khanna N, Gerull S, Buser A, et al. (2010) Evidence for a bidirectional relationship between cytomegalovirus replication and acute graft-versus-host disease. Biol Blood Marrow Transplant 16: 1309-1314.
- Sadeghi M, Daniel V, Naujokat C, Schnitzler P, Schmidt J, et al. (2008) Dysregulated cytokine responses during cytomegalovirus infection in renal transplant recipients. Transplantation 86: 275-285.
- Essa S, Pacsa A, Raghupathy R, Said T, Nampoory MR, et al. (2009) Low levels of Th1-type cytokines and increased levels of Th2-type cytokines in kidney transplant recipients with active cytomegalovirus infection. Transplant Proc 41: 1643-1647.
- Tishler M, Yaron I, Shirazi I, Yossipov Y, Yaron M (1999) Increased salivary interleukin-6 levels in patients with primary Sjögren's syndrome. Rheumatol Int 18: 125-127.
- Resende RG, Correia-Silva Jde F, Arão TC, Silva TA, Abreu MH, et al. (2010) Investigation of functional IL-10 gene polymorphism and IL-10 levels in acute graft-versus-host disease. J Clin Immunol 30: 465-473.









Table 1:
Baseline characteristics of patients and survival univariate analysis.
(n=63)
(n=25)
(n=38)