Authors:
Winfrida Cheriro1.,2, Gideon Kikuvi2, Simeon Mining3, Wilfred Emonyi3, Erick Rutto3, Elijah Songok4 and Michael Kiptoo4,5*
1Moi Teaching and Referral Hospital (MTRH), Kenya
2omo Kenyatta University of Agriculture and Technology (JKUAT), Kenya
3Moi University School of Medicine (MUSOM), Kenya
4Kenya Medical Research Institute (KEMRI), Kenya
5South Eastern Kenya University (SEKU), Kenya
Received:05 September, 2015;Accepted: 24 September, 2015; Published: 25 September, 2015
Prof. Michael Kiptoo, Principal Research Scientist, 5South Eastern Kenya University (SEKU), Tel: +254722756076; E-mail:
Cheriro W, Kikuvi G, Mining S, Emonyi W, Rutto E, et al. (2015) Drug Resistance Testing in HIV Infected Individuals on Treatment and Naive: Implications on Treatment Outcome. J HIV Clin Scientific Res 2(2): 062-068. DOI: 10.17352/2455-3786.000016
© 2015 Cheriro W, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
HIV/AIDS; Antiretroviral; Drug resistance; Drug resistance mutations
Background: The Government of Kenya started offering ART in the public sector since 2003. Despite the dramatic reduction in AIDS related morbidity and mortality, the emergence and spread of drug resistance (DR) threatens to negatively impact on treatment regimens and compromise efforts to control the epidemic. Therefore, there is a need for information on the situation of DR Mutations (DRMS) and their implications on treatment.
Objectives: To evaluate DRMS and their implications on treatment in HIV infected individuals attending Moi Teaching and Referral Hospital (MTRH) clinics.
Method: In 2009, we consecutively collected plasma samples from two groups of HIV infected individuals, antiretroviral (ARV) naive and ARV experienced for more than 12 months and failing therapy according to world health organisation (WHO) guidelines. We performed genotypic DR using well established in-house Sanger sequencing methods. We then followed up the patients and compared the DRMS in relation to their drug regimens at the time of sample collection and16 months later.
Result: We successfully extracted and sequenced 75 samples. Median age was 36.7 years. Out of 41 drug naive individuals only 3 had DRMS. Out of the 34 ARV experienced, 29 had DRMS to nucleoside reverse transcriptase inhibitor (NRTI), and 31 to non NRTI (NNRTI). After 16 months from sample collection date, 20/31(64%) ARV experienced patients with DRMS had not been changed therapy and only 5/20(25%) were susceptible to primary ARV while 12/14 changed were susceptible to new ARV.
Conclusion: The information obtained in our study can serve as an indicator of ARV program efficiency in patients still on treatment, those who are to start treatment and those who are to be changed therapy due to failure. DR testing would be necessary before initiating and /or changing ART in order to achieve optimal clinical outcome.
Abbreviations
AIDS: Acquired immunodeficiency syndrome; AMPATH: Academic Model Providing Access to Health care partnership clinics; ARDR: Antiretroviral Drug resistance; ART: Antiretroviral treatment; ARV: Antiretroviral; AZT: Zidovudine; CD4: Cluster of differentiation; DNA: Deoxyribonucleic acid; DR: Drug resistance; DRMs: Drug resistance mutations; EFV: Efavirenz; HAART: Highly active antiretroviral therapy; HIV: Human immunodeficiency virus; MTRH: Moi Teaching and Referral Hospital; NASCOP: National AIDS control program; NNRTI: Nucleoside reverse transcriptase; NRTI: Nucleotide reverse transcriptase; NVP: Nevirapine; PCR: Polymerase chain reaction; PI: Protease Inhibitors; PR: Protease; RT: Reverse transcriptase; SD:Standard deviation; 3TC: Lamivudine; TDF: Tenofovir; TDR: Transmitted Drug resistance mutations;
Introduction
Most data concerning Human Immunodeficiency Virus (HIV) non-B subtypes remain controversial. Highly Active Antiretroviral Therapy (HAART) has radically changed the clinical outcome of HIV, leading to decreased mortality and morbidity. Development of HIV drug resistance is inevitable in patients on ART. Increase in antiretroviral therapy (ART) in resource-limited settings (RLS) will successfully reduce HIV-related morbidity and mortality [1]. The increase in ART coverage is expected to lead to an increase in drug-resistant strains among experienced patients. Improved access to alternative combinations of antiretroviral drugs in sub-Saharan Africa is warranted [2].
As the rollout of ART in Kenya is on the rise, there is a need to monitor the patients on ART [3]. The use of Cluster of Differentiation (CD4) and viral load measurements is important in monitoring HIV patients both immunologically and virologically. Though virological and immunological monitoring is important, there is a need to provide HIVDR testing services for patients who are starting therapy and those who are suspected to be failing treatment before they are switched to a different regimen [4]. A public-health approach based on standardized, affordable drug regimens and limited laboratory monitoring is crucial in scale up efforts. The ever-expanding rollout of antiretroviral therapy in RLS without routine virological monitoring has been accompanied with development of drug resistance that has resulted in limited treatment success. A survey performed in Kampala showed a prevalence of transmitted drug resistance at 8.6% [5].
In Kenya, availability of ART is increasing. As ART use increases there is mounting evidence suggesting that DR will increase over time [6]. A recent cross-sectional study to determine treatment failure and drug resistance mutations among adults receiving first-line (3TC_d4T/AZT_NVP/EFV) and second-line (3TC/AZT/LPV/r) in Nairobi, Kenya, concluded that the detected accumulated resistance strains due to emergence of HIV drug resistance will continue to be a big challenge [7]. Another study carried out to evaluate treatment success and development of ART drug resistance at the Coast Province General Hospital, Mombasa, Kenya, revealed a high rate of treatment success after short term ART in patients treated at a public provincial hospital detected minority complex drug resistance profiles that were predictive of resistance to currently used second-line NRTIs and NNRTIs regimens [1]. In this article we present detailed data on DRMS from patients who had not started ARV and those who were failing with their implications on therapy. Identifying the relevant DRMS among non-B subtypes will be important for monitoring the evolution and transmission of drug resistance, determination of initial treatment strategies for persons infected with HIV non-subtype B [4]. According to International AIDS Society recommendations (IAS), evaluating susceptibility patterns among non-clade B persons should be a high priority because these viruses are by far the most prevalent world-wide. It is believed that surveillance will maximize the utility of first-line therapy and help minimize the cost of providing ART thereby sustaining current antiretroviral drug programs. The HIVDR testing is important as it gives the clinician accurate information of the most appropriate drug options. With ART scale up, there has been a need for monitoring for development of HIV drug resistance at a population level. Therefore, there is a need for country information on the situation of antiretroviral drug resistance (ARDR) to inform on policy guidelines.
Materials and Methods
Setting
The study was conducted at Moi Teaching and Referral Hospital (MTRH), AMPATH (Academic Model Providing Access To Health care partnership clinics), Eldoret, Kenya clinics.
The region includes the expansive Rift Valley, Western and Nyanza provinces, a cumulative population of about 15 million. The hospital is located in Eldoret Town in Uasin-Gishu (UG) County, which forms part of the UG Plateau West of the Great Rift Valley, at an altitude of 2118m above the sea level, latitude 00°30'52”N and longitude 035°17'52”E [2].
Study subjects
The study was conducted on isolates from patients who were known HIV positive attending the study site and met the selection criteria. During September 2009 and October 2011, patients receiving ARV therapy for at least 12 months and were suspected to be failing according to WHO guidelines were consecutively enrolled. After informed consent was obtained, a standardized questionnaire was administered to assess demographic, epidemiologic, clinical, and treatment information. ART-naive patients were also enrolled during the same period at the same study clinics. Samples from patients who had no history of exposure to ARV drugs and ARV drug naïve status according to a medical chart review and personal interview were collected consecutively.
Study design
The study conducted was a hospital-based prospective utilizing isolates from HIV positive patients who met the selection criteria. The study clinics provided ART according to the national guidelines for ART scale-up as recommended by WHO surveillance and monitoring surveys.
Data Collection Tools and Procedures
Laboratory procedures
Sample collection: Remnant blood samples collected for CD4 analysis from ARV naïve were centrifuged and plasma was collected. Remnant samples collected for viral load analysis from ARV experienced patients suspected to be failing therapy clinically were also centrifuged and plasma were collected and stored at −80°C.
HIV DNA extraction: HIV-1 nucleic acid was extracted from 400 µl of plasma using the Nuclisens Easy Mag system (Biomerieux, Canada) following manufacturer's instructions.
Reverse Transcription and polymerase chain reaction: HIV-1 protease (PR) and reverse transcriptase (RT) were bidirectional sequenced with an in-house protocol (8). Briefly, viral RNA was reverse transcribed and amplified according to the manufacturer's directions using the QIAGEN one-step RT-PCR kit (QIAGEN, Canada). The primers used were GaGp1-PR-out.for with a sequence of TGA ARG AIT GYA CTG ARA GRC AGG CTA AT and RT-new-out. Rev of CCT CIT TYT TGC ATA YTT YCC TGT T with nested primers GaGp6-PR-in.for YTC AGA RCA GRC CRG ARC CAA CAG C and RT-new-in.rev GGY TCT TGR TAA ATT TGR TAT GTC CA. All reactions were carried out using standard conditions using GeneAmp PCR System 9700 (ABI) thermocycler.
PCR product purification and sequencing
The PCR products were purified using Multi Screen Separations System as previously described [9] and diluted to 15 ng/ml for DNA sequencing. Amplicons were sequenced using ABI Prism Big Dye 3.1 Cycle Sequencing System (Applied Bio systems, USA) following manufacturer's instructions.
Data analysis
All the data generated in this study was saved in Microsoft Excel worksheets with a detailed database established to capture all the necessary information. Generated sequences were edited using Bio Edit v 7.0.5B. Aligned fasta files were uploaded to Stanford HIV Drug resistance (http://hivdb6.stanford.edu/asi/deployed/hiv_central.pl?program=hivdb&action=showSequenceForm). Phylogenetic relationships of newly derived viral sequences for comparisons with those of previously reported HIV group M from the Los Alamos database by CLUSTAL W profile alignment was utilized. To improve the accuracy of HIV-1 subtyping, the genotyping tool (http://www.ncbi.nih.gov/projects/genotyping/formpage.cgi) was used and the REGA sub typing tool (http://dbpartners.stanford.edu/RegaSubtyping/) was utilized as needed. Drug resistance mutation and subtype data collected from the Stanford HIV database sequence analysis program were manually input into appropriate excel spreadsheet file, verified and corrections made as needed. Categorical variables were presented in form of frequency tables while continuous variables were mainly summarized using means together with standard deviation and median.
To test significance of skewed continuous variables, Wilcoxon rank sum test was employed. Chi-square test was used to compare the association between categorical variables. Fisher's exact test was also used to compare categorical variables where some cells had expected value of less than 5. Level of significance was set at p < 0.05, with a 95 % confidence interval. All analyses were done using STATA version 11.0.
Ethical considerations
The study was approved by the Institutional Research and Ethics Committee (IREC) of the Moi University School of Medicine (MUSOM) and MTRH Review Board (IREC):(IREC/2010/06) and AMPATH (RES/STUD/17/2010).
Results
Socio-demographic characteristics
Among the 264 individuals who met the selection criteria, 128 were declared to be ART naïve and 136 had been on ARV for more than 12 months and were failing therapy clinically. Mean age was 37.01 years (SD=12.50). Majority were female 44(60.02%) (Table 1).
Polymerase Chain Reaction and Sequencing outcomes: One hundred and ten (110) samples were successfully amplified. Out of these, 75 samples were successful sequenced and analysed for the presence of drug resistance mutations. Out of 75, ARV experienced individuals failing therapy were 34 and most patients, 25 (73.5%) received 3TC + d4T/AZT + EFV/NVP as first-line treatment. Patients who reported treatment interruption or switch were 9(26.5%). Switch concerned mainly replacement of d4T or AZT by TDF or ABC and only 3(8.8%) had been switched to protease inhibitor (PI) regimens (Tables 1-3).
-
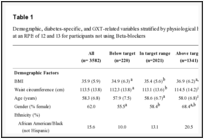
Table 2:
Patient Regimen.
-

Table 3:
Differences between Patient characteristics between the two study groups.
Drug resistance mutations in ARV naive
Drug resistance mutations were identified in 3/41 (7.3%) of patients and per drug class the values were as follows: 1 for PI and 2 for NRTI and 1 for NNRTI. In 1 patient, multiple mutations against 2 drug classes were seen, suggesting that they were probably not naïve (Table 4). Approximately 3/28(10.7%) of female subjects had DRMS. None of their male counterparts had mutations. There was no statistical difference when male vs. female respondents were compared (p=0.235).
-

Table 4:
DRMS in ARV Naive Patients.
Drug resistance mutations in ARV experienced
Among the ARV experienced patients who were failing therapy according to WHO guidelines, 34 samples which were successfully extracted and sequenced were studied. Mean age was 35.85years (SD=14.06). Majority were male 18(52.9%). Out of 34 samples, 27 had DRMS to nucleoside reverse transcriptase inhibitor (NRTI), 30 had DRMS to non-nucleoside reverse transcriptase inhibitor (NNRTI), and 2 had DRMS to NNRTI only (Table 5).
-

Table 5:
DRMS and level of resistance to baseline ARVs.
ARV therapy sixteen months after sample collection
Sixteen months after sample collection, 20/34 ARV experienced patients failing therapy were still on the same ARVs. Only 4/20(20%) of these patients were susceptible to the ARVs they were taking from DRMS analysis. Twelve out of the fourteen who were changed therapy were susceptible to the new drugs and 2 patients had their ARVs changed into other drugs that they were already resistant to (KE12-027, KE12043) (Table 6).
-

Table 6:
The ARV therapy, level of susceptibility and resistance.
Discussion
The results present depiction of the importance of HIV DR testing in a resource limited setting and their implications on treatment in both ARV naive and ARV experienced failing therapy before starting or changing therapy. However, we noted a low rate of amplification which may have been due to integrity of sample storage and transportation. Sequences obtained from 35 samples that were successfully amplified did not meet the integrity of good sequences for final drug resistance analysis.
Drug resistance mutations in ARV naive
In our study, drug resistant mutants were detected in three patients who were ARV naïve according to chart review. The prevalence of DRMs among drug naive populations revealed in our study might have been the result of the transmission of drug-resistant viruses from partners infected with the resistant virus or selection as a result of undisclosed use of ART. One patient had multiple NRTI drug resistance mutations, an indication that the patient may have had previous drug exposure. Although mutations conferring NRTI resistance have previously been reported among drug naive patients, the possibility that our patients had previous unreported contact with antiretroviral drugs could not be excluded [8].
Drug resistance mutations in ARV experienced
From our study, we observed that due to lack of information about any existing mutations before the therapy were changed, only 4/20(20%)who had not changed therapy were susceptible to the ARVs they were taking from DRMS analysis. Twelve out the fourteen who had their therapy changed were susceptible to the new drugs and the two patients had their therapy changed into ARVs they were already resistant to (KE12-027, KE12043). Most harboured a mutation at position M184I/V associated with 3TC and EFV resistance. The M184V on the other hand confer high level resistance to 3TC, a key backbone to first line antiretroviral treatment regimens in Kenya. The M184I mutation has been noted to be the first to appear but is quickly replaced by the M184V since this mutation has greater ability to induce higher replicative capacity [10].
Majority of the patients we reported with drug resistance mutations, were resistant to AZT and/or d4T because they harboured either mutation in the RT gene associated with resistance to RT inhibitors: multi NRTI-69 insertion complex b which affects all NRTIS (K70R (n=2), T215YF (n=5), K219QE (n=3). The presence of 3 of the following mutations M41L, D67N, L210W, T215Y/F, and K219Q/E has been associated with resistance to didanosin [6]. The presence of these mutations may improve subsequent virologic response to NNRTI-containing regimens (nevirapine or efavirenz) in NNRTI-naive individuals, although no clinical data exist for improved response to etravirine in NNRTI experienced individuals. KE12-027 had K65R, M184V while KE12043 had both M184V and T215Y. When associated with TAMs, M184V has been reported to increase abacavir resistance [11]. Studies have shown that the presence of K70R or M184V alone does not decrease virologic response to didanosine. The presence of mutations G190A had already compromised the use of next generation NNRTI etravirine. Importantly, presence of the K65R mutation compromises also the use of second-line regimens. Previous findings suggest that implementation of programs to consider the various socioeconomic and cultural barriers that may prevent successful uptake of antiretroviral prophylaxes are important [12].
Practical implications
Patients whose physicians have access to information about any existing mutations before the therapy are changed usually have more significant decreases in the viral load than patients in whom treatment is changed without knowledge of the resistance profile. Similar studies have led to development of new NRTIs, as well as new NNRTIs and PIs with different resistances profiles [3]. The options after treatment failures if improved will thereby increase the importance of resistance testing. On the other hand, simply changing national recommendations for initial ARV therapy from an NNRTI-based regimen to a protease inhibitor (PI)-based regimen would be suboptimal because PI-based regimens are more expensive and often less tolerated than NNRTI-based regimens. From our study, practical implications show that 80% (20/34) of patients whose therapy was not changed based on immunological and virological results only had DRMS to the regimens they were still taking while 85% (12/14) of those changed therapy were susceptible to the new regimens as per DRMS Stanford report. It is worth noting that M184V mutation is also associated with reduced viral replication and this may explain the reason for not changing regimen for the individuals that harbour this mutation.
Therefore, pre-therapy genotypic resistance testing would be useful to identify which patients should receive standard first-line therapy and which should receive a PI-containing regime [12].
Limitations
Viral load data was only available for ARV experienced patients who were failing therapy clinically.
Conclusion
The outcome of our study denotes high drug resistant strains in regions with non- B subtypes. The high prevalence of DRMs among drug experienced with evidence of drug failure populations revealed in our study might have been due to lack of DR analysis before start of therapy.
Acknowledgements
The authors would like to express their gratitude to the institutional research and ethics committee (IREC) and the AMPATH research office for permission to conduct this study. The authors also wish to thank the directors and staff of AMPATH clinics and laboratories, for providing logistics and technical support and for their cooperation.
Recommendations
Drug resistance testing would be necessary before initiating and /or changing ART in order to achieve an optimal clinical outcome. Similarly, there will be a need for a continued follow-up of persons with DRVS to determine clinical impact and help guide therapies for drug naïve and experienced with evidence of drug failure populations.
Authors' contributions
WC(MTRH) conceived the study, carried out field work, sample collection; sample analysis and manuscript preparation; GK(JKUAT) and SM(MUSOM) coordinated project implementation, WE(MUSOM) coordinated sample collection and analysis; ER(MUSOM) coordinated clinical oversight, ES(KEMRI) did project implementation oversight and overall supervision; MK(KEMRI/SEKU) coordinated field work, overall supervision and manuscript preparation. All authors read and approved the manuscript.
Sequence data
The sequence data has been deposited at Gen bank under accession numbers KP877888 to KP877961 and KR003380 to KR003405.
-
-
- Steegen K, Luchters S, Dauwe K, Reynaerts J, Mandaliya K, et al. (2009) Effectiveness of antiretroviral therapy and development of drug resistance in HIV-1 infected patients in Mombasa, Kenya. AIDS research and therapy 6: 12 .
- Einterz RM, Kimaiyo S, Mengech HN, Khwa-Otsyula BO, Esamai F, et al. (2007) Responding to the HIV pandemic: the power of an academic medical partnership. Acad Med 82: 812-818.
- Gilks CF, Crowley S, Ekpini R, Gove S, Perriens J, et al. (2006) The WHO public-health approach to antiretroviral treatment against HIV in resource-limited settings. Lancet 368: 505-510.
- Lwembe R, Ochieng W, Panikulam A, Mongoina CO, Palakudy T, et al. (2007) Anti-retroviral drug resistance-associated mutations among non-subtype B HIV-1-infected Kenyan children with treatment failure. J Med Virol 79: 865-872.
- Bennett DE, Camacho RJ, Otelea D, Kuritzkes DR, Fleury H, et al. (2009) Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: 2009 update. PloS one 4: e4724.
- Lihana RW, Khamadi SA, Lwembe RM, Kinyua JG, Muriuki JK, et al. (2009) HIV-1 subtype and viral tropism determination for evaluating antiretroviral therapy options: an analysis of archived Kenyan blood samples. BMC infectious diseases 9: 215 .
- Koigi P, Ngayo MO, Khamadi S, Ngugi C, Nyamache AK (2014) HIV type 1 drug resistance patterns among patients failing first and second line antiretroviral therapy in Nairobi, Kenya. BMC research notes 7: 890 .
- Lihana RW, Khamadi SA, Lubano K, Lwembe R, Kiptoo MK, et al. (2009) HIV type 1 subtype diversity and drug resistance among HIV type 1-infected Kenyan patients initiating antiretroviral therapy. AIDS res hum retroviruses 25: 1211-1217 .
- Ji H, Masse N, Tyler S, Liang B, Li Y, et al. (2010) HIV drug resistance surveillance using pooled pyrosequencing. PloS one 5: e9263 .
- Garcia-Lerma JG (2005) Diversity of thymidine analogue resistance genotypes among newly diagnosed HIV-1-infected persons. J Antimicrob Chemother 56: 265-269 .
- Johnson VA, Calvez V, Gunthard HF, Paredes R, Pillay D, et al. (2011) update of the drug resistance mutations in HIV-1. Top Antivir Med 19: 156-164 .
- Gupta RK, Goodall RL, Ranopa M, Kityo C, Munderi P, et al. (2014) High rate of HIV resuppression after viral failure on first-line antiretroviral therapy in the absence of switch to second-line therapy. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America 58: 1023-1026 .









Table 1:
Socio-demographic and clinical characteristics among study subjects.